Scientists have for the first time created ‘synthetic cells’ that can survive and reproduce on their own. A team lead by Dr Craig Venter published their results in the journal Science today.
Venter’s team set out to create a living cell based entirely on a computer-designed DNA sequence.
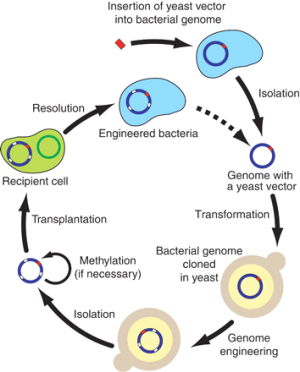
They digitally designed a stripped-down genome for the simple bacteria used in the experiment — much shorter than what occurs in nature, but still maintaining essential functions. From this computerised DNA sequence, a synthetic genome was chemically assembled, base-pair by base-pair, with the aid of yeast cells.
The scientists transferred the synthetic DNA to a living bacteria cell, which successfully reproduced. The new colony of ‘synthetic cells’ look and behave differently from the original, showing that the artificial genes have taken effect.
See below for a fact sheet on synthetic biology.
Dr Peter Dearden, Director of Genetics Otago comments:
“This paper represents a very significant step forward in engineering life. Venter’s group have been able to put a fully synthetic genome into a bacterial cell and get it to act as that cell’s genome. The technical skill required to get this to work is immense, and is the culmination of a long series of experiments that pioneered the technology to do this, as well as developing ways to prove that it had been done.
“The experiment raises an interesting question, has Venter and his team created life? The answer is no. Venter’s team is relying on the information in a natural genome. While the DNA strand that makes up the genome is synthetic and made in the lab, the information it contains comes from a species of bacterium; and it is the information that is important in a genome. Also Venter’s team needs a bacterial cell, one without a genome, to put their synthetic genome into. This cell, currently, can only be made by a living organism.
“However, while Venter and his team haven’t created life, they have carried out a remarkable feat, and put us one further step on the road to completely re-engineering organisms for biotechnological purposes.”
Dr Iain Lamont, Professor of Biochemistry at University of Otago comments:
“This is an amazing technical achievement that in years to come will be seen as a landmark in biology. It provides proof-of-principle that it is possible to make new microorganisms by synthesising their DNA and transplanting it into an existing cell, where it replaces the existing DNA. The very specialist technology allied to our ignorance of many aspects of biochemistry and physiology even in the simplest organisms means that we are very far from creating something completely new. Nonetheless, this work raises important ethical and philosophical questions that deserve wide discussion.”
Dr Neil Pickering, Senior Lecturer in Bioethics at University of Otago comments:
“Ethical issues arise both in questions about the nature of an activity and in questions about the consequences of an activity. And in both sorts of questions, the prospects for agreement on the answers are unclear.
“For instance, to describe the scientists in this case as ‘playing God’ is to characterise the nature of what they are doing. However, you may reject this characterisation. And even if you do accept it, depending on your view point, you may find this a fearful description or an invigorating one.
“The consequential issues that exercise people in such cases are of broadly two kinds: There are fears that the promised benefits to health, or the environment will be illusory, or outweighed by unintended effects – side effects if you like – of the use of novel organisms; and fears about possible evil intended effects e.g. from bioterrorism.
“The problems thinking through these consequential issues are many. But unless we outlaw an activity or allow it to go on unchecked, we have to face these problems. In particular, much is uncertain: what are the possible benefits and what the harms? What the likelihood of their coming about? There will be limitations on how effectively we can investigate these matters and reduce uncertainty.
“Moreover, appraising consequences cannot be an altogether objective activity. What to one may seem a risk worth taking for a benefit worth having may seem to another to be quite the opposite.”
Comments gathered from scientists by the UK Science Media Centre:
Dr. Gos Micklem, Department of Genetics at the University of Cambridge, said:
“This is undoubtedly a landmark paper. The group has been building towards this step and, from their earlier published work, are leaders at synthesising and re-assembling large segments of DNA. There is already a wealth of simple, cheap, powerful and mature techniques for genetically engineering a range of organisms. Therefore, for the time being, this approach is unlikely to supplant existing methods for genetic engineering. DNA synthesis is rapidly becoming cheaper and so this could change, but not soon.
“The technique could potentially come into its own if one wanted to introduce a large number of changes into an existing genome. However making a system that works predictably after introducing a large number of changes is one of the design challenges of the young field of synthetic biology: in the general case it is a challenge that is unlikely to be solved soon.”
Professor Richard Kitney OBE, Co-Director of the EPSRC Centre for Synthetic Biology at Imperial College London & Fellow of the Royal Academy of Engineering, said:
“An important aspect of the paper is that the original natural genome was sequenced and the sequence sent in alphanumeric form to a company call Blue Heron near Seattle. They then synthesised the sequence in 1000 bp cassettes and sent back the biological material to Venter et al, who reconstructed the entire genome and checked for errors.
“The synthesised genome was then placed in a different natural cell and replication took place. After 2 or 3 generations the second cell population took on the characteristics of the first population (i.e. a different bacterium). The paper has major implications for the harnessing of biology for industrial purposes.”
Professor Douglas Kell, Chief Executive of the UK Biotechnology and Biological Sciences Research Council (BBSRC), said:
“The ability to do all of the steps of protein synthesis from genome to product in order to make something that has a useful application is an important step in developing the potential of synthetic biology.
“Synthetic biology is a relatively new field and within the global research community, including in the UK, there is some truly avant-garde science happening. Together with EPSRC and Sciencewise-ERC, BBSRC has recently been exploring the range of perspectives of the UK public on synthetic biology to ensure that the cutting edge research that is carried out in this field is supported by policies that reflect the views, concerns and aspirations of the people who fund it – the UK taxpayers.
“As we become technically better at doing synthetic biology, the potential applications open up. Mainly, what it will allow us to do is harness useful biological processes so that we can make the sorts of products that are difficult to synthesise with traditional chemistry and physics.”
Professor Richard Kitney OBE, Co-Director of the EPSRC Centre for Synthetic Biology at Imperial College London & Fellow of the Royal Academy of Engineering, said:
“An important aspect of the paper is that the original natural genome was sequenced and the sequence sent in alphanumeric form to a company call Blue Heron near Seattle. They then synthesised the sequence in 1000 bp cassettes and sent back the biological material to Venter et al, who reconstructed the entire genome and checked for errors. The synthesised genome was then placed in a different natural cell and replication took place. After 2 or 3 generations the second cell population took on the characteristics of the first population (i.e. a different bacterium). The paper has major implications for the harnessing of biology for industrial purposes.”
Professor Paul Freemont, Co-Director of the EPSRC Centre for Synthetic Biology at Imperial College London, said:
“The paper published in Science today by Craig Venter and colleagues is a landmark study that represents a major advance in synthetic biology. Venter and colleagues have for the first time demonstrated that a single genome of around 1 million base pairs can be chemically synthesised and assembled correctly and transplanted into a recipient cell.
“The step change advance, which has alluded them in previous publications, is that they have now demonstrated that the transplanted synthetic DNA can be ‘booted up’ to operate the functions of the new recipient cell in terms of replication and growth. Although the recipient cell is not man-made but is another natural cell, what Venter’s team have shown is that after transplantation and multiple cell divisions the recipient cell take son the characteristics or phenotype of the newly transplanted genome. (This is like taking a Mac computer operating systems and installing it onto a PC and the PC becoming a Mac computer.)
“This is a remarkable advance as it now provides a ‘proof of concept’ that we can chemically synthesise and assemble full genomes and transplant them into recipient cells, which after selection contain only the synthetic genome, and after rounds of cell division become a new and one might argue synthetic cell. The applications of this enabling technology are enormous and one might argue this is a key step in the industrialisation of synthetic biology leading to a new era of biotechnology.
“Of course one also needs to be cautious, as it is not clear if this approach will work for larger and more complex genomes or for transplantation in different bacterial cells. However, this is a landmark step in our abilities to manufacture man-made cells for man-made purposes.”
Additional information from Professor Freemont:
“In detail the paper describes the chemical synthesis and assembly of the 1.08Mbp genome of Mycoplasmamycoides. This organism is a small bacteria and lives as a parasite in cattle and goats. Mycoplasma lack cell walls, have no discernable shape and are the smallest (0.1 µM) known free-living life forms and are most likely to have evolved from Gram-positive bacteria. They are present in both animal and plant kingdoms and act as colonisers.
“The choice of Mycoplasma by the Venter group for genome synthesis and transplantation is based on the small size of the genome and for the mycoides species has a reasonably fast growth rate. The difficulties they report in terms of getting the synthetic genome booted up were due to a single base pair mutation in an essential gene (dnaA) which they noted after several attempts. Correcting this mutation allowed the synthetic genome to work properly, although its not clear how the mutation occurred – whether in the synthesis or assembly step. The transplantation process involves using an antibiotic selection process where the newly transplanted genomes infer resistance to the transplanted cell to live in the presence of a lethal antibiotic.
Science Media Centre Fact Sheet
(Provided by the UK Science Media Centre)
Synthetic Biology
What is synthetic biology?
The creation or modification of living biological systems through artificial means, drawing on natural biological principles and engineering techniques. This involves creating and manipulating biological components at the cellular and genetic level, with the ultimate aim of creating simple artificial organisms (‘artificial life’).
Is it a new area of research?
The term “synthetic biology” first appeared in the literature in 1978, to describe bacteria that had been genetically engineered using recombinant DNA technology. In this respect, synthetic biology was largely synonymous with genetic engineering, though this has since changed.
In 2000, the term “synthetic biology” was first used to describe the synthesis of unnatural organic molecules that function in living systems. More broadly in this sense, the term has been used with reference to efforts to “redesign life”.
(Ref: Benner, S. et al. “Synthetic Biology”, Nature Reviews Genetics, July 2005)
Is synthetic biology research underway in New Zealand?
According to the Ministry of Research, Science and Technology: “Currently there is no significant synthetic biology research activity in New Zealand. While most important developments will occur in other countries, fundamental research is moving relatively quickly and aspects may be taken up here.
“While genetic material in the form of DNA or RNA is routinely imported into the country with limited oversight of the type and nature of the information it contains (re)creation of a synthetic organism will be covered by the Hazardous Substances and New Organisms Act since it involves in vitro genetic modification. The debate is currently limited in New Zealand and the potential human and environmental impacts of synthetic biology are yet to be put forth for wider debate or considered in a regulatory sense.
“MoRST held a ‘Bioissues Forum’ for government policy makers on this topic in April 2007 to alert government agencies to potential challenges that developments in synthetic biology may raise.”
What are the techniques?
The main techniques involved are genetic sequencing and synthesis of simple bacterial genomes – one of the primary aims is to construct a synthetic genome with the minimum number of genes necessary to support life – and then transplanting this into a bacterial cell, thereby creating a new type of organism.
Another key aspect of synthetic biology is the creation of biological parts, devices and systems which do not occur in the natural world, as well as redesigning existing systems. For example, biological parts can be used to produce biologically-based devices, such as biosensors for urinary tract infections.
Scientists at the J. Craig Venter Institute in the US (led by Craig Venter) are attempting to do this; so far they have successfully created an artificial genome but not yet (prior to the current paper) succeeded in integrating it into a bacterial cell.
What are the applications?
- Synthesis of pharmaceuticals – synthetic organisms can be genetically ‘programmed’ to synthesise pharmaceutical compounds – e.g. the synthesis of the anti-malarial compound artemisinin by introducing novel metabolic enzymes into yeast (Ro et al., Nature 440; 2006)
- Living therapies – synthetic organisms can also be designed to carry out therapeutic functions – e.g. the use of modified bacteria to specifically target tumour cells in cancer therapy (Zhao et al. PNAS 2005). Although this example involves creating transgenic bacteria without using engineering principles, development of such therapies will be expanded and speeded up using synthetic biology approaches.
- Engineering human stem cells to have controlled outputs and characteristics – applications might include tissue regeneration or manipulation of the immune system to treat specific infections and diseaese
- Microbial biosensors – The fields of biomaterials and microbial biosensors are well established, but are moving rapidly into adopting synthetic biology approaches – e.g. University of Edinburgh’s IGEM 2006 project entry, which aimed to create an arsenic biosensor that could work in parts of the world most affected by poor water quality (e.g. Bangladesh). Also Imperial College London’s entry in 2007 on biosensors for bacterial infections.
- Biological logic gates – the biological equivalents to a key component of computers and microprocessors – the aim is to be able to produce biological microprocessors which could control intracellular functions.
-
New energy sources – e.g. biofuels such as sugar cane and maize are inefficient as much of the plant material is cellulose, which cannot be broken down easily. Synthetic micro-organisms could be developed to break down the cellulose.
-
New materials – synthetic cells could be programmed to produce artificial materials with wide applications – e.g. the lab of US scientist Chris Voigt is working on engineering elements of Salmonella bacteria to secrete spider silk proteins, for possible use in artificial biomaterials.
-
Non-biological devices that incorporate biological ‘components’
-
Is it right to synthesise life?
-
Bioterrorism – developing new viruses/ recreating extinct viruses (e.g. 1918 influenza virus). However, some terrorism experts believe this method of attack is unlikely.
- Biohackers – As genetic information multiplies and the cost of hardware falls, could we see a new generation of “biohackers” akin to computer hackers? This is unlikely as regulation exists making it difficult to obtain key DNA sequences and very secure biofacilities are required for such research.
-
Accidental leakage of new pathogens into the environment – However, scientists should be able to build in “self-destruction” mechanisms that prevent the new organism from replicating out of control.
Info from the International Genetically Engineered Machines Competition:
Royal Academy of Engineering report on synthetic biology:
This is a fact sheet issued by the Science Media Centre to provide background information on science topics relevant to breaking news stories. This is not intended as the ‘last word’ on a subject, but rather a summary of the basics and a pointer towards sources of more detailed information. These fact sheets are checked by one or more experts in the field and can be read as supplements to our ’round-up’ press releases.